A Reference Guide to Deep Learning in Pytorch | Tips and Tricks
Every now and then I will come across a tensor manipulation method in Pytorch or some piece of code in Pytorch and think “I wish I had known about this earlier”. Then after one or two days completely forget about that thing. This article is a reference for such useful Pytorch code snippets.
File Structure Organization
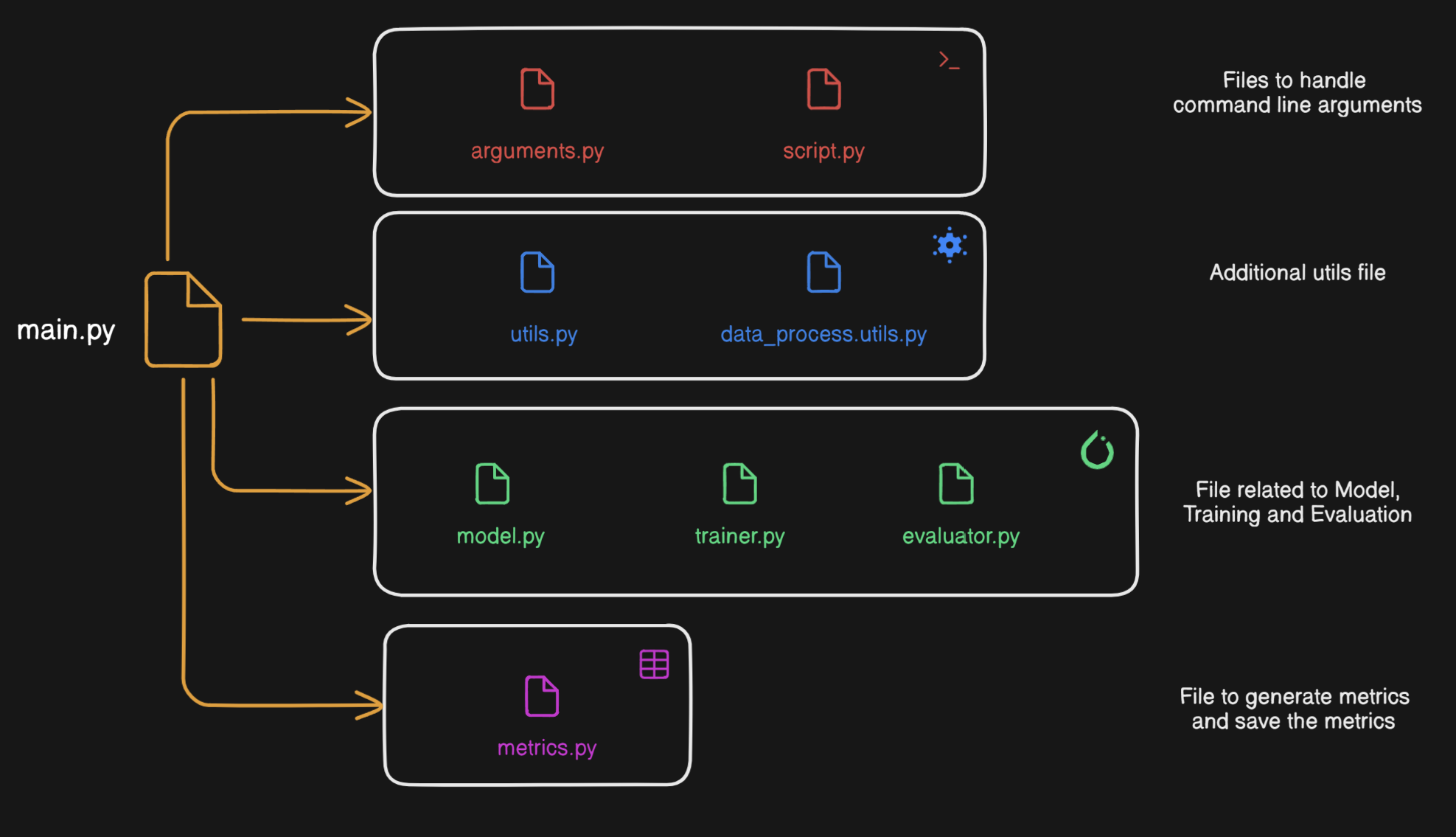
File structure organization depends on the complexity of the project and personal preference. For small to medium project I more or less use the following file structure.
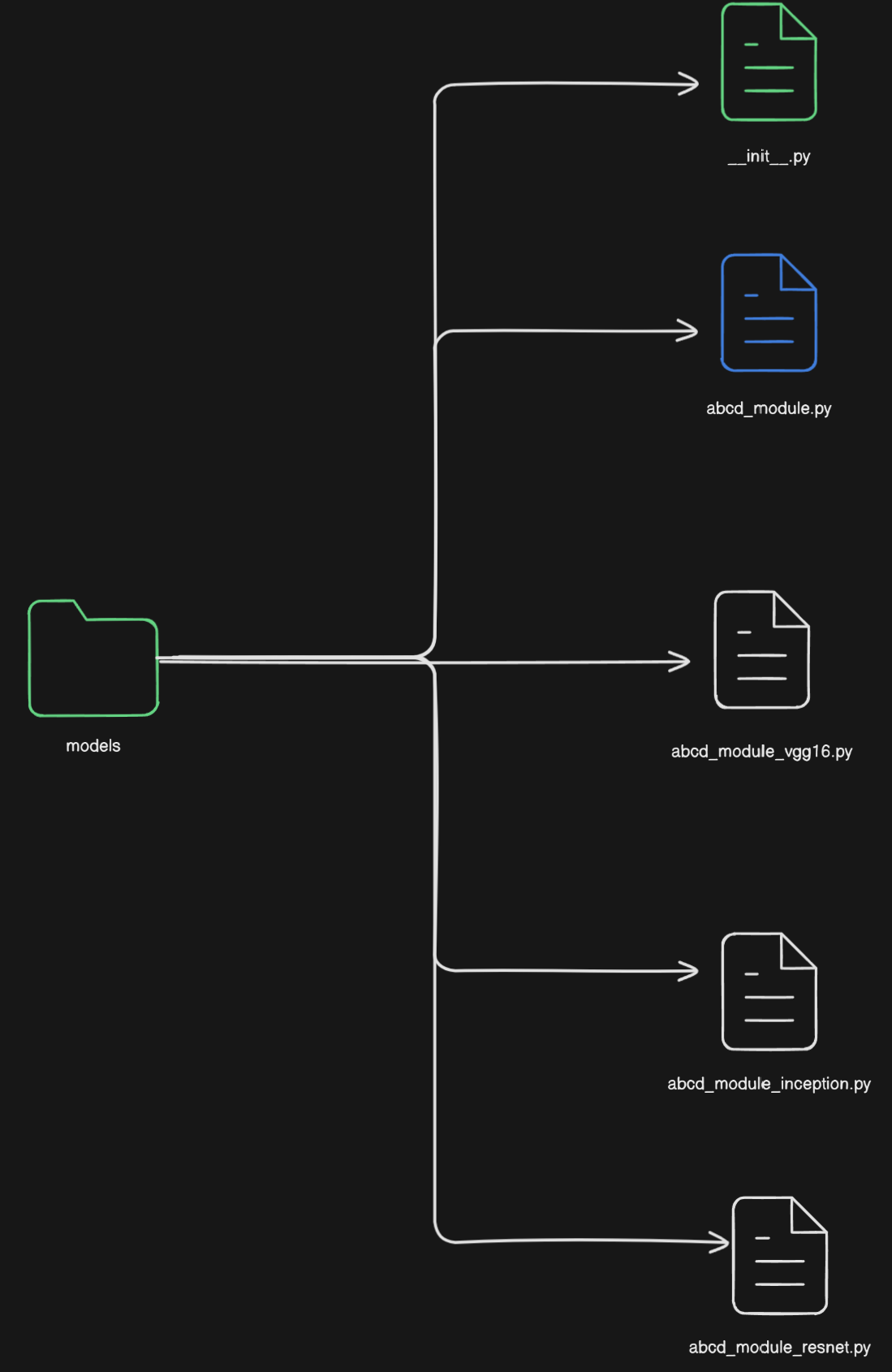
It might be the case that, the model.py file is so complex you need to break it down into multiple files. Another scenario might be you need to run your code against different architectures. In that case, there should be a folder called models and inside that folder there should be different .py files for different architectures.
As a hypothetical scenario imagine, you are proposing a new block called “abcd_module”. And you want to check how the module behaves with different architectures like ResNet, VGG-16, InceptionNet, etc. Then your folder structure should look like above. The __init__.py should define how you will import the different files.
Note that this should be done if your architectures defined in those files are radically different from each other. If the difference is couple of layers depending on a bunch of choices, you are better off with a single model.py and using ModuleDict or ModuleList combined with if-elif-else blocks.
Also, another disclaimer you should only move to a multi-file appproach for your architecture if you model is too complex. If it is a simple model breaking it down into multiple files will cause more annoyance.
Code organization
Saving command line arguments to a JSON file
I use argparser to process command line arguments and use a script.py file to get the command line arguments. Example code snipppet from one of the project I used this:
script.py
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
import subprocess
command = [
'python', 'main.py',
'--folder','Exp1',
'--epoch','50',
'--split_ratio','0.8',
'--seed','66',
'--batch_size', '32',
'--dim_input','6',
'--hidden_emb_size','32',
'--learning_rate','0.001',
'--weight_decay','1e-3',
'--dropout','0.2',
'--frac_graph_to_backdoor','0.4'
]
subprocess.run(command)
As the first argument I need to specify the main file and the second arugment is the folder where I want to save everything from a specific experiment.
arguments.py
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
import argparse
import json
import os
class parse_args():
"""
- Creates Arguments Parser object
- Creates a dictory with the folder name specified by --folder argument
- Creates a json file with the same name as the folder and stores the arguments
"""
def __init__(self):
argParser = argparse.ArgumentParser(description='arguments')
argParser.add_argument('--folder', type=str,
help='Name of the folder that will contain related info and results from a specific experiment')
argParser.add_argument('--epoch', type=int,
help='Training epoch count')
argParser.add_argument('--split_ratio', type=float,
help='Train-Test Split Ratio')
argParser.add_argument('--seed', type=int,
help='Random seed value for reproducibility')
argParser.add_argument('--batch_size', type=int,
help='Batch Size to be used for DataLoader')
argParser.add_argument('--dim_input', type=int,
help='Dimension of the input feature vector of each node')
argParser.add_argument('--hidden_emb_size', type=int,
help='Dimension of the feature vectors in the hidden layers')
argParser.add_argument('--learning_rate', type=float,
help='Learning Rate for Training')
argParser.add_argument('--weight_decay', type=float,
help='Weight Decay Parameter')
argParser.add_argument('--dropout', type=float,
help='Dropout Value')
argParser.add_argument('--frac_graph_to_backdoor',type=float,
help='Fraction of Training Graphs that need to be backdoored')
self.args = argParser.parse_args()
def get_args(self):
return self.args
def dump_json(self):
folder_name = self.args.folder
json_file=folder_name # json file name with the same name as the folder
os.makedirs(folder_name, exist_ok=True) # Create folder if it doesn't exist
json_path = os.path.join(folder_name, self.args.folder)
with open(json_path, 'w') as json_file:
json.dump(vars(self.args), json_file, indent=4)
You will notice I am creating a json file with the same name as the folder and storing the command line arguments in there.
In the main.py I just need to do the following:
1
2
3
4
5
6
from arguments import parse_args
argument_parser=parse_args()
argument_parser.dump_json()
args=argument_parser.get_args()
Separate Trainer class to train model
I usually write the training portion of the code in main.py file. But a much better approach is to have a separate trainer class to train the model. You can have additional functions like store_values to store different metrics and outputs (for example embeddings) from the model. You can also provide an additional variable like train_log=['True',20] which will decide if you should log the training metrics and how frequent the training logs should be. For some model training, it does not make sense to log metrics after every epoch. Finally, you can return the trained model in the train function.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
class Trainer():
def __init__(self,model,data,optimizer,num_epochs,train_log=['True',20]):
self.model=model
self.data=data
self.optimizer=optimizer
self.num_epochs=num_epochs
self.train_log=train_log
self.losses = []
self.accuracies = []
self.outputs = []
def accuracy(self,pred_y, y):
return ((pred_y == y).sum() / len(y)).item()
def store_values(self,loss,acc,out):
self.losses.append(loss)
self.accuracies.append(acc)
self.outputs.append(out)
def train(self):
if self.train_log[0]:
print('Training Info:')
print('-------------')
for epoch in range(self.num_epochs):
self.model.train()
self.optimizer.zero_grad()
out = self.model(self.data.x,self.data.edge_index)
loss = F.nll_loss(out[self.data.train_mask], self.data.y[self.data.train_mask])
acc = self.accuracy(out[self.data.train_mask].argmax(dim=1), self.data.y[self.data.train_mask])
self.store_values(loss,acc,out)
if self.train_log[0]:
if epoch % self.train_log[1] == 0:
print(f'Epoch: {epoch} | Training Loss: {loss} | Training Accuracy : {acc} ')
loss.backward()
self.optimizer.step()
return self.model
Tensor Manipulation Methods
torch.Tensor.new_ones()
Creates a new tensor but with all elements initialized to the value 1. By default, the returned Tensor has the same
torch.dtypeandtorch.deviceas the original tensor.
1
2
tensor = torch.tensor((), dtype=torch.int32)
tensor.new_ones((2, 3))
torch.unbind()
Removes a tensor dimension. Returns a tuple of all slices along a given dimension, already without it.
1
2
3
4
torch.unbind(torch.tensor([ [1, 2, 3],
[4, 5, 6],
[7, 8, 9]]))
1
(tensor([1, 2, 3]), tensor([4, 5, 6]), tensor([7, 8, 9]))
torch.Tensor.tolist()
Returns the tensor as a (nested) list.
1
2
a = torch.randn(2, 2)
a.tolist()
1
2
[[0.012766935862600803, 0.5415473580360413],
[-0.08909505605697632, 0.7729271650314331]]
tensor1.expand_as(tensor2)
Expands the dimensions of a tensor to match the shape of another tensor.